Biomarker Predictors of Omalizumab Efficacy in Asthma
A recent publication by Djukanović et al. demonstrates the promise that VOC breath biomarkers have to predict the monocloncal drug Omalizumab clinical efficacy in severe asthma
| Publication information: Djukanović R, Brinkman P, Kolmert J, Gomez C, Schofield J, Brandsma J, et al. Biomarker Predictors of Clinical Efficacy of the Anti-IgE Biologic, Omalizumab, in Severe Asthma in Adults: Results of the SoMOSA Study. Am J Respir Crit Care Med. 2024 Apr 18. DOI: 10.1164/rccm.202310-1730OC
Disease Area: Asthma Sample medium: Breath, blood, induced sputum, and urine Analysis approach: Multi-omic approach: GC-MS, eNose, FeNO, LC-MS, UHPSFC-IM-MS/MS, LC/HDMSE, etc. Summary:
|
Introduction
Diagnostic methods for asthma are unable to predict who will respond better to different treatment options, meaning the identification of an appropriate treatment for asthma is based on a ‘trial and error’ approach. This involves treatments being prescribed sequentially and monitored for response over time, resulting in unnecessary spending for healthcare systems, and periods of poor disease control with an increased risk of exacerbations for the patient. One such treatment is the anti-IgE monoclonal antibody omalizumab which can significantly reduce asthma exacerbations, however, there is no way to predict which asthma patients will benefit from this. New biomarkers with the ability to predict which patients would benefit from omalizumab therapy in advance would hasten the process, and remove the need for trial and error. Therefore, the authors of this study utilized an innovative multi-omic approach in those with severe asthma to assess the potential links between responsiveness to omalizumab and the composition of exhaled breath, blood, induced sputum, and urine.
Methods
Participants with severe asthma were included in this study (open-label, real-world design), all of whom received standard-of-care omalizumab. The study was conducted in two phases. For phase 1, patients were evaluated by physician’s assessment (the Global Evaluation of Treatment Effectiveness – GETE tool) after 16 weeks of treatment early responses. For phase 2, patients were evaluated at 52 weeks for ≥50% decrease in asthma exacerbations or dose of maintenance oral prednisolone (mOCS).
A total of 191 patients provided exhaled breath, blood, induced sputum, and morning urine samples before and after 16 weeks of treatment. To test for late-responders, 173 of these participants were also assessed for improved clinical symptoms at 52 weeks. Four analytical omics methods were applied: including both Gas Chromatography-Mass Spectrometry (GC-MS) and eNose technology to analyze exhaled breath samples. These were then compared with more common clinical biomarkers: blood and sputum eosinophil count, exhaled nitric oxide through FeNo breath testing, and with an evaluation tool based on GETE.
Results
The multi-omic analysis resulted in a total of 1408 variables which were captured across the sampling mediums utilized, including 70 breath VOCs, and 158 eNose signatures without molecular identities. VOC breath analysis generated two panels of 5 VOCs, one for predicting early improvement and one for predicting > 50% exacerbation reduction. Urine compounds, physician assessment (GETE), blood/ sputum eosinophil counts, and serum IgE could not predict whether omalizumab would reduce exacerbation reduction.
Analysis of all the omics platforms demonstrated that breath VOCs from GC-MS (rather than eNose) and blood lipidomics had the highest ability to predict both early and late responses (Figure 4B and C from Djukanović et al. – inserted below). All other omics platforms assessed had weak predictive ability, and therefore were not considered potential candidate biomarkers. There were two panels of five exhaled breath VOCs which demonstrated potential value as biomarker panels. The first panel consisted of benzothiazole, acetophenone, 2-pentylfuran, methylene chloride, and 2-methylbutane, which could predict early improvement with a Receiver Operating Characteristic Area Under the Curve (ROCAUC) of 0.835. The second panel of VOCs were 2-ethyl-1-hexanol, toluene, 2-pentene, nonanal, and a VOC of unknown identity, which could predict ≥50% exacerbation reduction with an ROCAUC of 0.780.
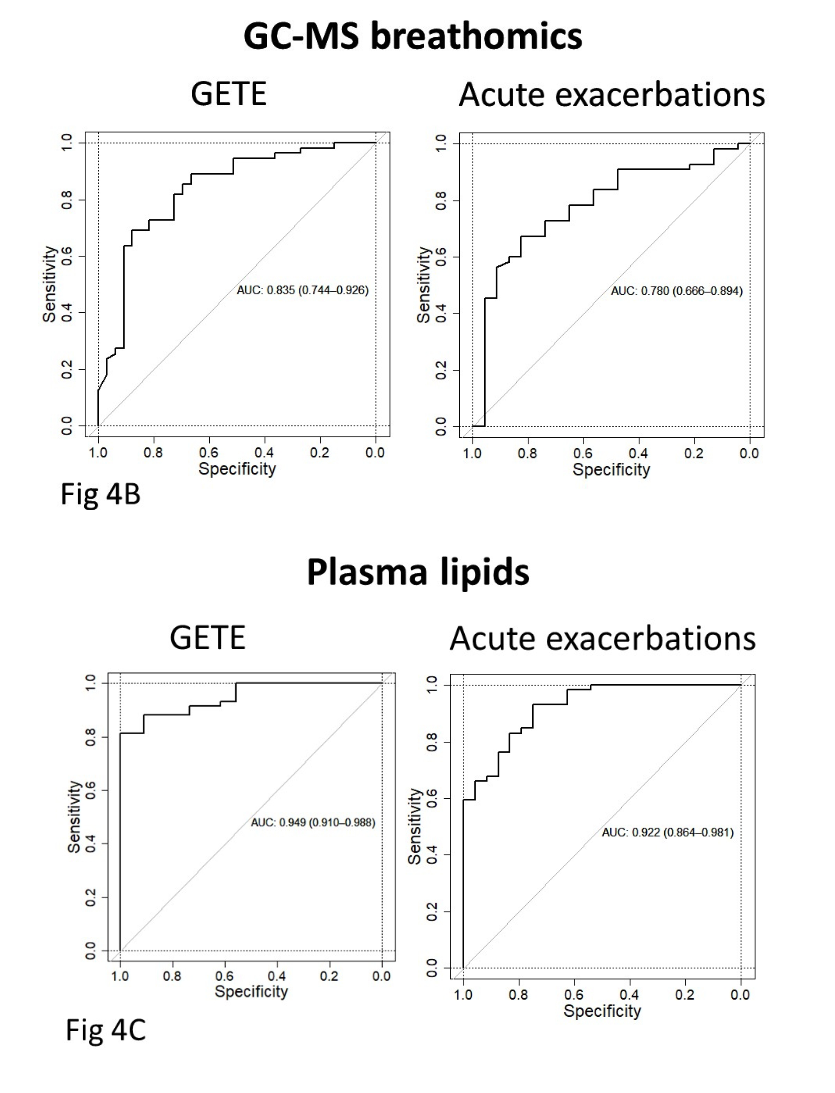
Figure 4 (from Djukanović et al. 2024). Receiver operating characteristic area under the curve (ROCAUC) diagrams for GC-MS breath VOC analysis (breathomics) (4B), and plasmic lipidomics (4C) are shown.
Discussion
This study used a multi-omic approach to evaluate the predictive performance of thousands of potential biomarkers at one time for response to the monoclonal antibody therapy omalizumab in severe asthmatics. The superior platforms for predictive biomarkers for responsiveness were shown to be VOC breath analysis, and lipids from blood analysis. Five breath VOCs (2-ethyl-1-hexanol, toluene, 2-pentene, nonanal, and one unknown VOC), confidently predicted the reductions in exacerbations, and another set of five breath VOCs (benzothiazole, acetophenone, 2-pentylfuran, methylene chloride, and 2-methylbutane) could predict early responses. Together, these VOCs demonstrated the ability to differentiate between mild/moderate asthmatics and all atopic severe asthmatics (ROC AUC 0.931), and between mild/moderate asthma and severe asthmatics prone to exacerbations (≥2 exacerbations per annum). This is meaningful as this is a cut-off for initiating treatment with a biologic such as omalizumab. Some of these identified breath VOCs have been previously associated with asthma, lung inflammation previously (1,2), and have known mechanisms of production, for example, from lipid peroxidation from oxidative damage (3,4). Many are also present in our Breath Biopsy® VOC Atlas, and can be confidently detected using our Breath Biopsy® OMNI® method.
Although lipids had greater predictive ability than breath VOC biomarkers, the authors of this study considered breath VOC analysis to be a superior biomarker discovery and analytical platform due to several key attributes. This included the ease of breath collection, and an easier development pathway of point-of-care devices for use in a clinical setting. GC-MS-based approaches were seen to outperform eNOSE-based approaches, as an eNose approach could not predict clinical improvement. This suggests that ability to assign VOC identities, rather than broad overall patterns is more informative for clinical use, and can pull out a greater depth of detail for superior biomarker development.
This study demonstrates that breath VOCs are a powerful, yet currently underutilized sampling medium as part of biomarker discovery studies. VOCs can originate from endogenous metabolic processes impacted by disease, such as lipid peroxidation resulting from inflammatory states. In this study, several VOCs could be grouped together for greater predictive ability, a result that reflects other studies in the literature that show that groups of VOCs perform better as a biomarker group as opposed to any single VOC. If unique patterns of VOCs in the breath can be linked to clinically valuable information, such as predicting the response to drug therapies, or diagnosis, breath biomarkers could be developed into next-generation non-invasive clinical tests. This study also demonstrates the potential for combining the analysis of several sampling mediums together to achieve more powerful biomarker performance. The key compounds identified from breath VOC analysis and blood lipid analysis were chemically distinct, and yet together could provide greater sensitivity and specificity for clinical application.
Breath collection and analysis is challenging due to the nature of breath being comprised of both VOCs that have interacted with underlying physiology (both endogenous and exogenous VOCs), and those that have been inhaled immediately prior to breath sampling from the background air. This landscape requires highly sensitive analytical procedures that can confidently detect and measure compounds at very low concentration, and rigorous quality control procedures to ensure that genuine signals are distinguishable. We have the expertise to implement breath VOC analysis, providing both breath sampling collection hardware, analysis, and biological interpretation of VOCs in context of what is known about their mechanistic origin based on internal data and the broader literature. This includes state-of-the-art high-resolution accurate mass TD-GC-MS, and internal reference libraries to support high-confidence compound identification for better standardization and ultimate translatability of identified biomarkers.
Get in contact with us to discuss how breath VOC analysis could complement your multi-omic studies and provide an alternative biomarker platform for discovery.
References
- Ibrahim W, Wilde MJ, Cordell RL, Richardson M, Salman D, Free RC, et al. Visualization of exhaled breath metabolites reveals distinct diagnostic signatures for acute cardiorespiratory breathlessness. Science Translational Medicine. 2022 Nov 16;14(671):eabl5849. DOI: 10.1126/scitranslmed.abl5849
- Zanella D, Henket M, Schleich F, Dejong T, Louis R, Focant JF, et al. Comparison of the effect of chemically and biologically induced inflammation on the volatile metabolite production of lung epithelial cells by GC×GC-TOFMS. Analyst. 2020 Jul 29;145(15):5148–57. DOI: 10.1039/D0AN00720J
- Drabińska N, Flynn C, Ratcliffe N, Belluomo I, Myridakis A, Gould O, et al. A literature survey of all volatiles from healthy human breath and bodily fluids: the human volatilome. J Breath Res. 2021 Apr 21;15(3). DOI: 10.1088/1752-7163/abf1d0
- Ratcliffe N, Wieczorek T, Drabińska N, Gould O, Osborne A, Costello BDL. A mechanistic study and review of volatile products from peroxidation of unsaturated fatty acids: an aid to understanding the origins of volatile organic compounds from the human body. J Breath Res. 2020 May;14(3):034001. DOI: 10.1088/1752-7163/ab7f9d