Detection of COVID-19 in breath aerosols
Exhaled breath aerosols are less sensitive than swabs but may relate to transmission
| Publication information: Smolinska et al. The SARS-CoV-2 viral load in COVID-19 patients is lower on face mask filters than on nasopharyngeal swabs, Scientific Reports (2021) DOI: 10.1038/s41598-021-92665-3
Disease Area: Infectious diseases (COVID-19) Application: Early detection, Transmission control Sample medium: Breath Products: Face masks Analysis approach: PCR Summary:
|
The COVID-19 pandemic has sparked an enormous amount of new research, with over 160,000 papers mentioning it currently listed in PubMed. Notably, it has led to increased interest in breath sampling as a means to detect and diagnose illnesses, and various approaches have been explored to use either exhaled breath condensate (EBC), exhaled breath aerosols (EBAs), or volatile organic compounds (VOCs) to identify infection.
EBAs consist of respiratory droplets of a range of sizes which are a key vector for SARS-CoV-2 transmission. The ability to capture EBAs and analyze them to detect virus particles represents a significant opportunity to improve virus detection using non-invasive methods.
Aerosol generation and limit of detection
In 2020, Owlstone Medical® initiated a collaboration to investigate the efficacy of using filters in face masks, such as those in the ReCIVA® Breath Sampler, to capture EBAs for analysis using established PCR workflows [1]. Initially, we used inactivated viruses, both SARS-CoV-2 and HCoV-NL63, to investigate the limit of detection for samples collected on filters (Figure 1). The system used produced EBAs across a range of sizes, from 0.2 to 5 μm. The limit of detection for both virus types was shown to be less than 10 copies per filter, suggesting that our approach is highly sensitive for virus detection.
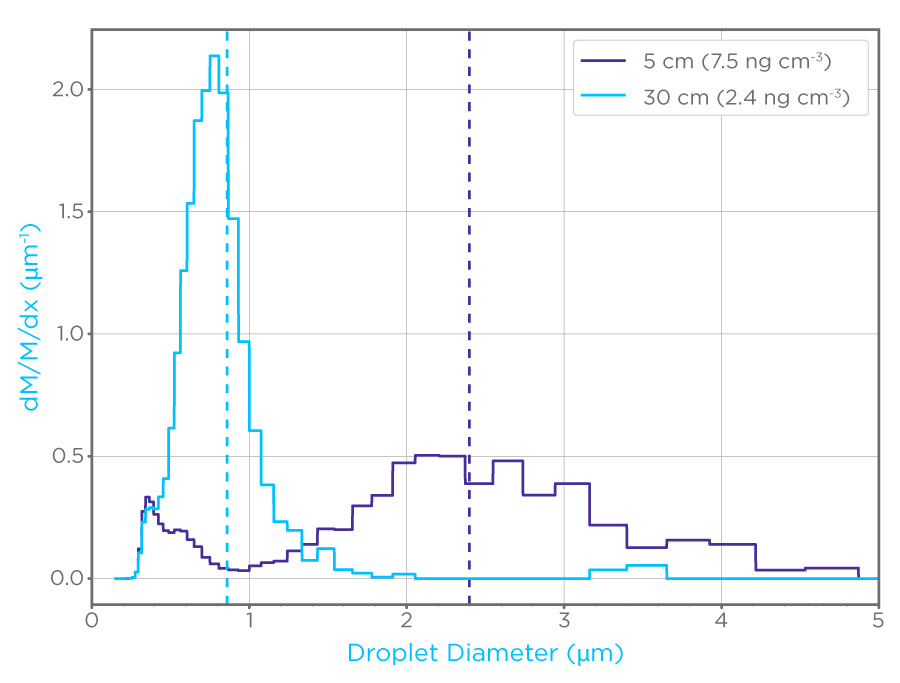
Figure 1: Particle probability density by mass (dM/m/dX) of aerosols generated to mimic EBAs, note the difference in distribution between the 5cm dilution path (purple) and the 30 cm dilution path (cyan). These aerosols were used to assess the limit of detection for coronaviruses on filters.
COVID-19 detection on face masks
Subsequently, 47 hospitalized COVID-19 patients were tested using the filters to see if they could be used to detect the infection in vivo. Patients had previously received a positive nasopharyngeal swab result before being admitted to hospital but had variable times between the initial test and the subsequent breath collection, ranging from a single day to more than a week. Of the 47 patients, only four gave positive results based on breath samples (8.5% sensitivity; Figure 2).
Thirty nine of the patients had a second swab test at the same time as breath collection. Of these, three were found to no longer be infected. Of the remaining 36, two had positive results based on breath sampling (5.6% sensitivity).
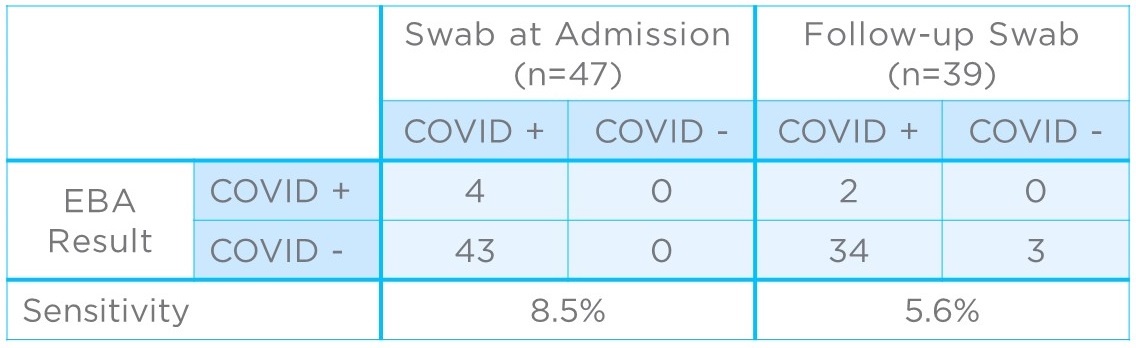
Figure 2: Clinical results comparing COVID-19 detection from EBAs to nasopharyngeal swabs collected at admission (left) and, where possible, as a follow-up to EBA collection (right).
Analysis
The laboratory-based results show that filters can be effective at trapping virus particles from EBAs for high sensitivity detection via PCR. Yet, despite this sensitivity to virus detection, EBAs appeared to show very low sensitivity for clinical detection of COVID-19 infection except for patients tested at the earliest stages of the disease cycle. The majority of patients produced undetectable levels of SARS-CoV-2 particles on breath.
What these results may suggest is that the majority of patients studied were not, at the time, producing infectious aerosols, and may not have been capable of transmitting the infection to others. It could also be the case that only some patients ever produce high numbers of airborne viral particles and that these are super spreaders that contribute the most to virus transmission. This is supported by previous studies on rhinovirus [2].
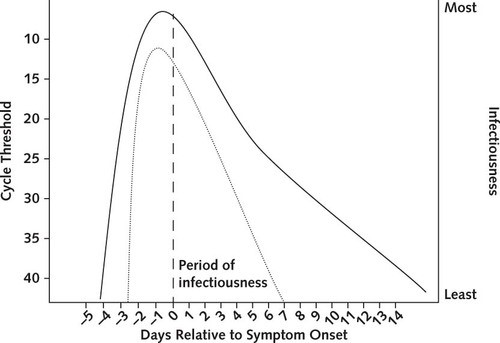
Figure 3: Infectiousness and cycle threshold profiles for COVID-19 infection. Cycle threshold is inversely related to the number of viruses in a sample, and is lowest prior to symptom onset. (Meyerowitz et al. 2021) [3]
An independent study from Zhang et al. found similar results in an experimental study of COVID-19 infected cynomolgus monkeys (Macaca fascicularis) [4]. A key finding here was that virus particles on breath were only detectable for the first six days post-infection and peaked after just two days post-infection. While there are likely to be differences in the infection profile between macaques and humans, this result contributes to the view that the majority of patients in Smolinska et al. may have reached the stage of infection where they no longer released significant numbers of viral particles on breath.
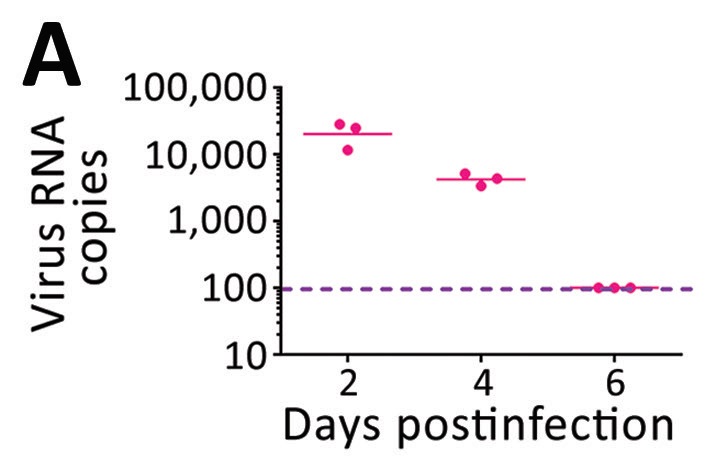
Figure 4: Data from cynomolgus monkeys showing declining numbers of exhaled COVID-19 viruses as infection progresses. (Zhang et al. 2021) [4]
Compared to the results of Smolinska et al., other published studies that used face masks to capture EBAs from COVID-19 positive patients, reported around 40% positive PCR results. Notably, these studies consistently collected EBA samples between 24 and 36 hours after the initial nasopharyngeal swab, which may explain the significantly higher positive detection rate. This is further supported by unpublished results based on longitudinal sampling which show that viral load detected from EBA increases before it is detectable via swabs but falls rapidly compared to other sampling methods.
Summary
While EBA collection is highly sensitive for airborne virus detection, it is not an effective tool for detecting later stages of COVID-19 infection. However, the relationship between viral shedding via EBA and virus transmission may mean that EBA collection via face masks could be developed as a tool to assess the risk of transmission between individuals. This is supported by several other similar studies that show that transmission risk is highest during the early stages of infection but falls long before the infection is cleared by the immune system.
We have also published a press release discussing the results of this study.
References
- Pubmed search accessed July 2021 https://pubmed.ncbi.nlm.nih.gov/?term=COVID-19&sort_order=asc&size=50&show_snippets=off
- Smolinska, A., et al., The SARS-CoV-2 viral load in COVID-19 patients is lower on face mask filters than on nasopharyngeal swabs. Sci Rep, 2021. 11(1): p. 13476. https://doi.org/10.1038/s41598-021-92665-3
- Fabian, P., et al., Origin of exhaled breath particles from healthy and human rhinovirus-infected subjects. J Aerosol Med Pulm Drug Deliv, 2011. 24(3): p. 137-47. https://dx.doi.org/10.1089%2Fjamp.2010.0815
- Meyerowitz, E.A., et al., Transmission of SARS-CoV-2: A Review of Viral, Host, and Environmental Factors. Ann Intern Med, 2021. 174(1): p. 69-79. https://doi.org/10.7326/M20-5008
- Zhang, C., et al., SARS-CoV-2 Aerosol Exhaled by Experimentally Infected Cynomolgus Monkeys. Emerg Infect Dis, 2021. 27(7): p. 1979-1981. https://doi.org/10.3201/eid2707.203948
Quick Start Guide: Everything you need to know about how breath analysis can be used in infectious disease research
