Mass spectrometry imaging and direct analysis
Direct, real time analysis of molecular information from biological samples
Direct analysis of samples using mass spectrometry is now possible using ambient ionization techniques such as DART (direct analysis in real time), DESI (desorption electrospray ionization) and LESA (liquid extraction surface analysis).
DESI and LESA can be employed when analyzing samples on substrates such as thin tissue sections, bacterial cultures or dried blood spots. They also allow in situ, real time analysis of spatial molecular information from samples, allowing a picture of chemical composition to be formed. Increasingly, direct analysis is finding applications in cancer diagnosis, as well as in the characterization of lipids, proteins and metabolites.
The chemical complexity of natural samples provides an inherent challenge in the application of direct analysis techniques, which prevents comprehensive detection and characterization of molecular species. Isobaric interferences in the mass spectrum of a sample complicate tandem MS analysis for structural characterization and obscure visualization of an ion’s spatial distribution.
Our chip-based ion mobility system, ultraFAIMS, can facilitate direct analysis by acting as a tuneable filter that rapidly separates gas phase ions at atmospheric pressure based on differences in their mobilities in an oscillating electric field. This allows selective transmission of subsets of ions or classes of molecules, which is particularly useful in imaging applications where the spatial distribution of particular compounds in a sample are being mapped.
 |
Adding ultraFAIMS to your direct analysis workflow helps to:
If you’re interested in using ultraFAIMS ion mobility in a direct analysis or mass spectrometry imaging application, we’ve compiled an ebook including papers, presentations and posters that showcase how researchers are using the system to improve their results. |
Ambient Ionization and FAIMS Mass Spectrometry for Enhanced Imaging of Multiply Charged Molecular Ions in Biological Tissues
Clara L. Feider, Natalia Elizondo and Livia S. Eberlin
In this study, issues relating to sample complexity in direct analysis are overcome using ultraFAIMS separation prior to mass spectrometric analysis. The system was able to reduce chemical noise and to improve signal-to-noise ratios, sensitivity, and dynamic range.
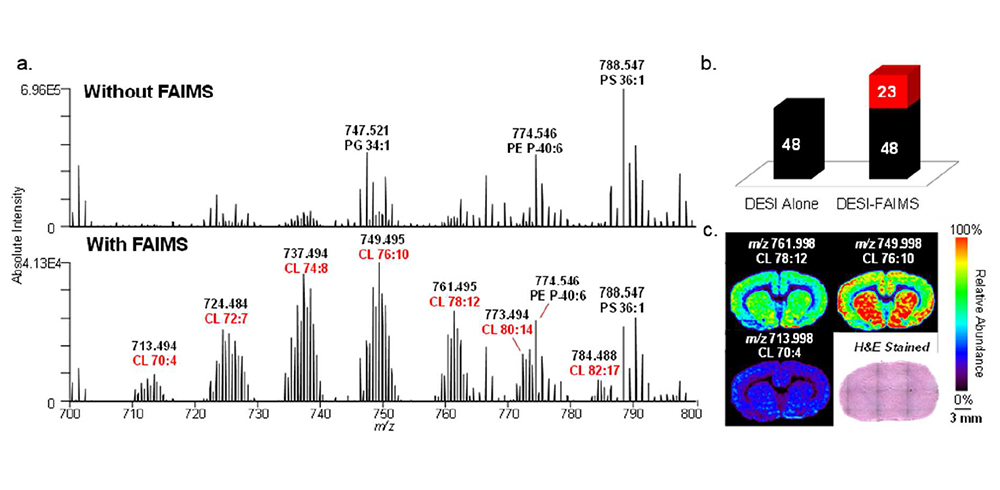
ultraFAIMS was integrated with both DESI-MS (desorption electrospray ionization – mass spectrometry) and LMJ-SSP-MS (liquid-microjunction surface sampling probe – mass spectrometry) to image and characterize metabolites, glycerophospholipids, glycosphingolipids, and multiply-charged proteins in rat brain, human thyroid, and human ovarian cancer tissues (see example figures).
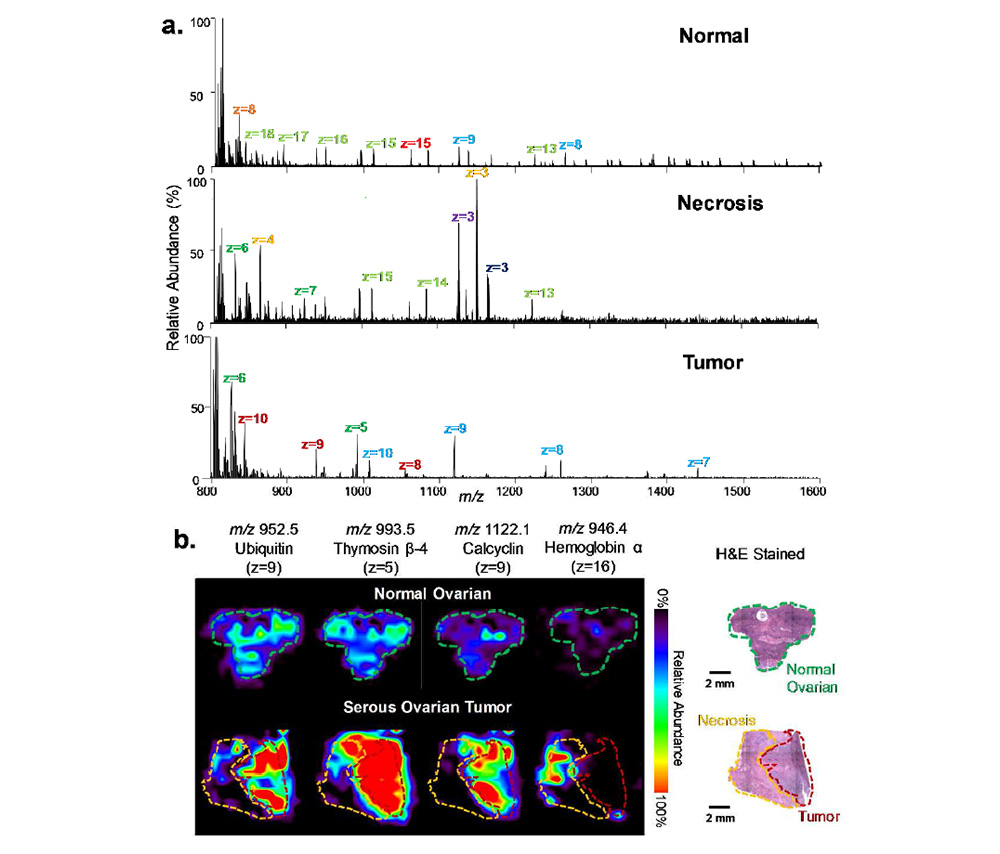
Using this approach, the authors show the first example of global protein imaging in human cancerous tissue by ambient ionization mass spectrometry imaging. The results indicate that integration of ultraFAIMS with DESI or LMJ-SSP is extremely valuable for imaging selected molecular classes in biological tissues.


