Identifying pathogens on breath via the lung microbiome
Earlier diagnosis of microbial strains allows for more effective therapies
| Publication information: Waqar Ahmed et al. Microbial volatiles as diagnostic biomarkers of bacterial lung infection in mechanically ventilated patients, Clin. Infect. Dis. 2022, Oct 31:ciac859. DOI: 10.1093/cid/ciac859
Disease Area: Respiratory disease (Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae and Escherichia coli) Application: Early detection Sample medium: Breath Analysis approach: GC-MS Summary:
|
Ventilator associated pneumonia (VAP) is the most common secondary infection in critically ill hospitalized patients. Pneumonia is a broad term for the symptoms caused by a number of pathogens – ideally rapid identification of the actual strain would allow prescription of targeted anti-microbial therapy, the most effective treatment. However, current techniques for determining the causative pathogen can be invasive and time consuming and any delay in treatment is associated with worse patient outcomes. Unfortunately, the broad-spectrum antimicrobial therapies that are prescribed instead can lead to drug-induced toxicity,microbiome dysbiosis, and contributes to the emergence of antimicrobial resistant strains of pathogens.
New techniques to rapidly identify causative pathogens in suspected cases VAP will facilitate timely and effective clinical decision making. Previous studies have reported changes in the volatile organic compounds (VOCs) on breath of infected patients and suggested that the microbial volatilome could differ between strains – meaning that early detection of causative strains could be possible using breath VOCs. That breath testing is both non-invasive and possible while a patient is still ventilated makes this option even more attractive.
Ahmed et al. have carried out a translational study attempting to compare and unify results from both in vitro and in vivo work in order establish strong prospective biomarkers of a number of pathogenic strains associated with VAP (Staphylococcus aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae and Escherichia coli).
Method
Bacterial strains were cultured in nutrient broth and volatile products of microbial metabolism were captured using a headspace sampler. Breath samples were collected from 89 ventilated patients within 24 hours of clinical suspicion of VAP, by sampling ventilator circuit tubing into sorbent tubes. Both bacterial headspace and patient breath VOCs were then analyzed using thermal desorption-gas chromatography-mass spectrometry (TD-GC-MS).
Patients also provided bronchoalveolar lavage (BAL) cultures which were used to identify the specific pathogens present in their lungs. Approximately half of the sample group (44 patients) did not develop pneumonia and were pathogen negative via BAL.
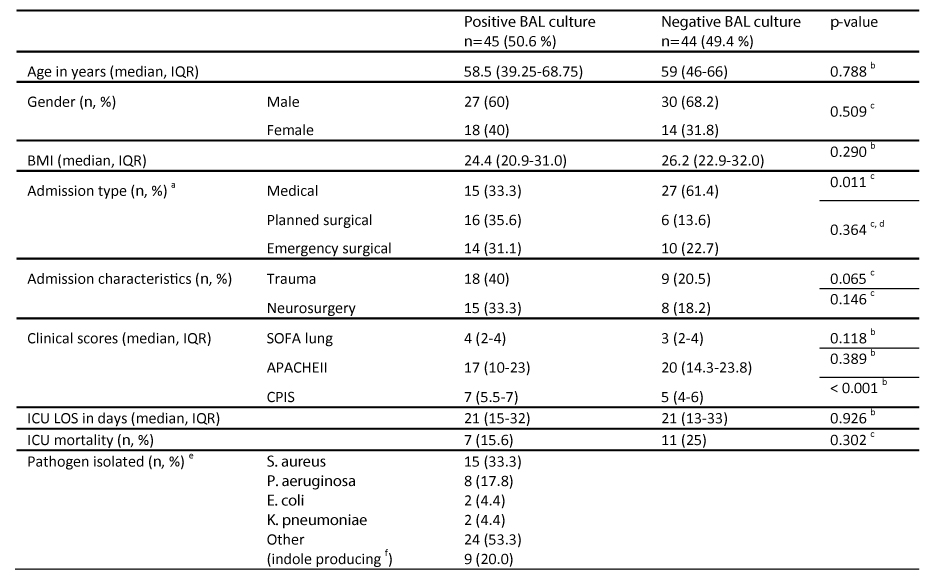
Table 1: Patient clinical characteristics
The analysis data set contained over 1000 molecular features (MFs). These MFs were analyzed to identify VOCs in breath samples that could be attributed to individual bacterial strains.
Results
A total of 19 VOCs were detected from the in vitro pathogen cultures after VOCs present on controls had been removed from the data set. Certain VOCs were unique to particular pathogens. These included 1-undecene, methyl thiocyanate, dimethyl sulfide, and 2-aminoacetophenone, which were only captured from P. aeruginosa cultures; ethyl acetate, 2-heptanone, and 2-nonanone which were only captured from K. pneumoniae cultures; and benzaldehyde and indole which were only found to be produced by the E. coli cultures.
These 19 microbial VOCs were then compared to the patient breath sample data, and five were eliminated from further analysis because they could not be confidently identified on breath. For P. aeruginosa, acetone and 3-methylbutanal were found to be lower than in other pathogen-positive samples. For S. aureus, 3-methylbutanal and 3-methylbutanoic acid concentrations were found to be significantly higher than both pathogen-negative and other pathogen-positive samples.
Some VOCs showed differentiation between pathogen positive and pathogen negative patient samples. For the S. aureus-positive samples, 3-methylbutanal and 3-methylbutanoic acid concentrations showed a weak level of correlation with each other and were assessed for their diagnostic potential against both the culture-negative controls and the other pathogen-positive groups. The 3-methylbutanal-based model produced an AUROC of 0.87 against controls and 0.81 against other pathogen positive groups, performing marginally better than a 3-methylbutanoic acid-based model in both cases.
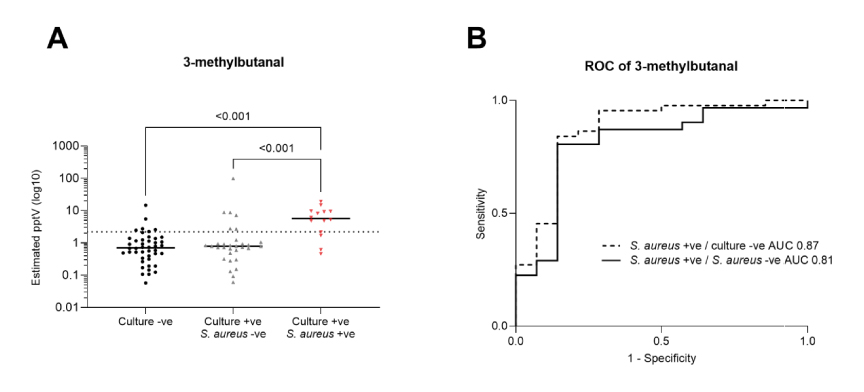
Figure 1: Scatter plots of estimated VOC concentrations and ROC curves of 3-methylbutanal. Scatter plots are annotated with the group median (black line), and estimated limit of detection (dotted line).
Discussion
Both methylbutanal and 3-methylbutanoic have previously been associated with S. aureus in published reports in both in vitro and in vivo studies, and this work confirms that connection. Their correlation suggests they may share a microbial metabolic pathway of origin, possibly leucine metabolism.
The effects of active medical interventions in these patients remains an area for further investigation. Although breath samples were collected before antibiotic treatment for VAP had begun, the patients participating in this study were critically ill and may have been administered additional medications that would be reflected in their VOC profiles. Further work and larger studies are needed validate these findings and investigate any potential confounding impacts of additional therapies administered to patients breathing with the aid of ventilators.
This study has demonstrated the strength of using a translational approach to investigate prospective microbial biomarkers, showing the potential for non-invasive breath tests to diagnose pathogens. At Owlstone Medical we have found in vitro analysis an extremely helpful way to accelerate our progress in breath test development, particularly in the areas of lung cancer and liver disease, to date.
Whether you’re interested in the human metabolome, or the microbiome, we can support your in vitro and in vivo breath studies through Breath Biopsy® OMNI® – the most advanced solution for reliable global breath VOC analysis.
