VOC biomarkers indicative of Eosinophilic Inflammation in Asthma
A recent publication by Peltrini et al. discovered 19 VOCs associated with eosinophilic inflammation in asthma, and subsequently validated this finding in exhaled breath in two independent cohorts
| Publication information: Peltrini R, Cordell RL, Wilde M, Abuhelal S, Quek E, Zounemat-Kermani N, Ibrahim W, Richardson M, Brinkman P, Schleich F, Stefanuto PH. Discovery and Validation of a Volatile Signature of Eosinophilic Airway Inflammation in Asthma. American Journal of Respiratory and Critical Care Medicine. 2024 May 31(ja). DOI: 10.1164/rccm.202310-1759OC.
Disease Area: Asthma Sample medium: Sputum headspace, breath Analysis approach: TD-GC-MS Summary:
|
Introduction
Fractional exhaled nitric oxide (FeNo) breath tests measure exhaled nitric oxide (NO) as a marker of airway inflammation for the assessment of asthma and are currently used in the clinic. However, there is a need for better biomarkers to improve the accuracy of this test as it can be modified by corticosteroid use, as well as other confounders like diet, smoking, and the presence of rhinitis (irritation/ inflammation in the nose) (1), as well as additional breath tests for a wider number of respiratory conditions. Although all asthma patients experience a common set of symptoms, the underlying disease mechanisms are different between patients, and this impacts which treatment is best. Understanding the phenotypes responsible for asthma in each patient is key to improving treatment. The inflammatory process of asthma can be categorized into two major types based on the predominant type of immune cells causing the inflammation: eosinophilic (type 2) or neutrophilic (type 1). Most asthmatic patients have type 2 inflammation.
No further breath VOC tests for asthma have yet come to market, despite the extensive literature and previous work (2,3). Better methods of biomarker discovery are needed, and better standardized approaches and robust methodology (4–6). This study sought to combine the power of headspace VOC analysis of sputum from patients with severe asthma to first identify candidate biomarkers of the underlying processes of asthma (eosinophilia), and then identify these VOCs in a targeted study in two independent cohorts for validation.
Methods
N = 36 adult patients with severe asthma were recruited, with inclusion criteria into the study including at least six weeks after exacerbation, and on stable treatment for at least 4 weeks. Sputum samples were collected from all patients, and 1L of headspace emitted from these samples was collected onto sorbent tubes. The sputum samples were categorized into either eosinophil enriched (n=22) or non-eosinophil enriched (n=14) based on a 3% threshold upon differential cell counting. Headspace and control samples (background room air headspace) were analyzed using TD-GC-MS. A validation study of identified VOCs from headspace sputum analysis used exhaled breath from two separate cohorts from EMBER (n=65) and U-BIOPRED (n=42). Breath from the EMBER cohort was collected using the ReCIVA® Breath Sampler and analyzed using GC×GC-FID-MS, and Tedlar bags from the U-BIOPRED cohort, which were analyzed using GC-MS. FeNO data was available for most of the U-BIOPRED cohort only, however, blood or serum eosinophil was available for at least some participants in both.
Results
Sputum headspace analysis
Analysis of the volatiles emitted directly from the sputum samples detected 393 features after thresholding (based on the frequency of observation) and the removal of artifacts. An elastic net regression analysis was used to identify VOCs associated with sputum eosinophilia ≥3%, which resulted in the selection of 19 VOCs. These 19 VOCs included five aldehydes: hexanal, nonanal, decanal, benzaldehyde, 2-ethylhexanal, three alkanes: tridecane, decane, cyclohexane isothiocyanate, two alcohols: 1-hexanol, 2-butoxy-ethanol, one halogen: methylene chloride, one ketone: acetone, and seven aromatic hydrocarbons: p-xylene, benzene, toluene, phenol, styrene, α-methylstyrene, and benzothiazole (Figure 1). This biomarker signature could discriminate between non-eosinophil-enriched sputum AUC of 0.90 [0.80-0.99] (p<0.0001), demonstrating a greater ability than blood eosinophils (AUC: 0.61 [0.42-0.80]) or FeNO (AUC:0.6 [0.35-0.86]).
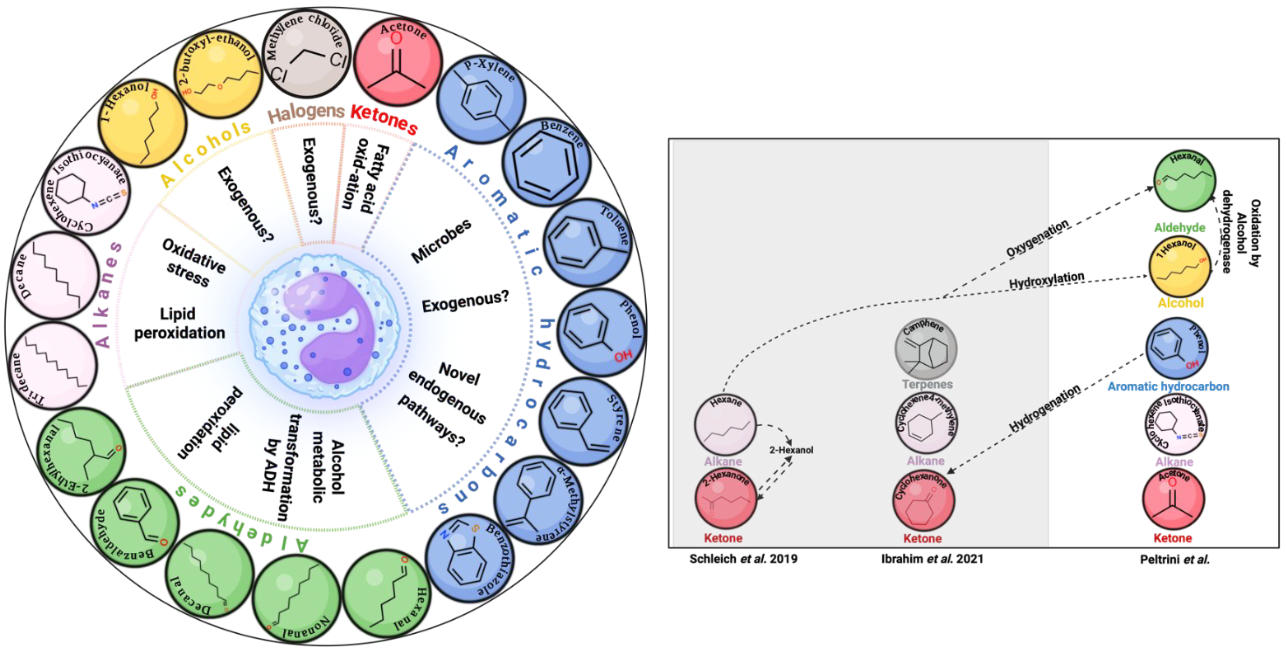
Figure 1 – Image and caption from Peltrini et al., 2024. (Left hand panel) An illustration of discovered eosinophilic breath volatile biomarker, their chemical structures and the metabolic pathways that could potentially be related to each chemical group. (Right hand panel) previously identified breath biomarkers in severe asthma studies characterizing patients according to sputum eosinophilia and arranged to show the possible chemical relationship and similarities with the reported discovery biomarkers in this study.
Clinical validation
Exhaled breath from both the EMBER and U-BIOPRED cohorts were analyzed. In the EMBER cohort, 16 of the 19 VOCs identified from the headspace analysis were detectable in the breath, the missing three VOCs being phenol, 2-butoxyethanol, and benzothiazole. In the U-BIOPRED cohort, 17 of the 19 headspace VOCs were detectable in the breath, the missing VOCs this time were 1-hexanol and 2-butoxyethanol. Ultimately, the 19 VOCs demonstrated good diagnostic accuracies for detecting sputum eosinophilia in both the EMBER and U-BIOPRED cohorts (AUROCs: ~0.90), with combined sensitivities and specificities of >0.80.
Discussion
The findings of this study identified candidate VOC biomarkers are associated with sputum eosinophilia with high sensitivity and specificity, and strongly support the potential of breath VOC biomarkers for clinical use. The study design utilized a robust discovery pipeline with VOC headspace analysis and external validation in two independent disease cohorts. All of the 19 VOCs categorized as a VOC biomarker signal of eosinophilic airway inflammation in asthma were identified using MSI level 1 criteria, previously highlighted as a key goal for improving the standardization of breath research during the Ask the Expert panel at BBCon 2023 (5). All of the VOCs identified in this study have also previously been identified as being associated with asthma, highlighting the validity and reliability of the data presented. Interestingly, the authors note that oxygenation, hydroxylation, or hydrogenation of several of the reported VOCs produce biomarkers that have previously been associated with sputum eosinophilia (7–9).
In the discovery cohort and U-BIOPRED, the VOC signature was shown to significantly outperform FeNO, the only currently available breath test that is used for clinical investigation for asthma.
In this study, several VOCs could be grouped together for greater predictive ability, a result that reflects other studies, including another recent study in those with severe asthma we previously covered that showed that groups of VOCs perform better as a biomarker group as opposed to any single VOC (12). The VOCs in this study were divided into two panels of breath VOCs that could predict responses to omalizumab, of which three VOCs were common to both this and the previous study: toluene, nonanal, and benzothiazole. Phenol was identified as part of the 19 VOCs associated with eosinophilia, however, this VOC is commonly associated with contamination from sampling hardware such as bags (10,11). Phenol was able to be removed from the data without affecting the diagnostic accuracy of the breath VOC signature, with an AUROC value of 0.90 (CI 95% 0.75-1.0).
The VOCs associated with eosinophilic inflammation have now been reliably found and identified as biomarkers of asthma that can be studied in future work. Analysis of the headspace of biological samples allows for greater control over experimental conditions to collect more in-depth, mechanistic data about the origin of VOCs in the breath of the disease area of interest, to better understand the underlying biology for clinical interpretation. The headspace analysis to validation in breath is powerful for the VOC biomarker discovery framework utilized as part of this study, and we can offer this capability through our headspace analysis pipeline, OMNI platform for complete breath collection and analysis pipeline, and the ReCIVA Breath Sampler used in this study. To find out more information about breath biomarkers for asthma, and other potential research studies involving the use of VOCs, please do not hesitate to contact us.
References
1. Chan EY, Ng DK, Chan C hong. Measuring FeNO in Asthma: Coexisting Allergic Rhinitis and Severity of Atopy as Confounding Factors. Am J Respir Crit Care Med. 2009 Aug;180(3):281–281. DOI: 10.1164/ajrccm.180.3.281
2. Azim A, Barber C, Dennison P, Riley J, Howarth P. Exhaled volatile organic compounds in adult asthma: a systematic review. European Respiratory Journal. 2019 Sep 1;54(3). DOI: 10.1183/13993003.00056-2019
3. Ibrahim W, Wilde MJ, Cordell RL, Richardson M, Salman D, Free RC, et al. Visualization of exhaled breath metabolites reveals distinct diagnostic signatures for acute cardiorespiratory breathlessness. Science Translational Medicine. 2022 Nov 16;14(671):eabl5849. DOI: 10.1126/scitranslmed.abl5849
4. Herbig J, Beauchamp J. Towards standardization in the analysis of breath gas volatiles. J Breath Res. 2014 Sep;8(3):037101. DOI: 10.1088/1752-7155/8/3/037101
5. Chou H, Godbeer L, Ball M. Establishing Breath as a Biomarker Platform – Take Home Messages from the Breath Biopsy Conference 2023. J Breath Res. 2024; DOI: 10.1088/1752-7163/ad3fdf
6. Lawson JLD, Nakhleh MK, Smolinska A. Reproducibility and reporting, the routes to progress in breath research—highlights from the Breath Biopsy Conference 2021. J Breath Res. 2022 Jun;16(3):030401. DOI: 10.1088/1752-7163/ac661d
7. Peltrini R, Cordell RL, Wilde M, Abuhelal S, Quek E, Zounemat-Kermani N, et al. Discovery and Validation of a Volatile Signature of Eosinophilic Airway Inflammation in Asthma. American Journal of Respiratory and Critical Care Medicine. 2024 May 31 ; DOI: epdf/10.1164/rccm.202310-1759OC
8. Schleich FN, Zanella D, Stefanuto PH, Bessonov K, Smolinska A, Dallinga JW, et al. Exhaled Volatile Organic Compounds Are Able to Discriminate between Neutrophilic and Eosinophilic Asthma. Am J Respir Crit Care Med. 2019 Aug 15;200(4):444–53. DOI: 10.1164/rccm.201811-2210OC
9. Ibrahim B, Basanta M, Cadden P, Singh D, Douce D, Woodcock A, et al. Non-invasive phenotyping using exhaled volatile organic compounds in asthma. Thorax. 2011 Sep;66(9):804–9. DOI: 10.1136/thx.2010.156695
10. Steeghs MML, Cristescu SM, Harren FJM. The suitability of Tedlar bags for breath sampling in medical diagnostic research. Physiol Meas. 2007 Jan;28(1):73–84. DOI: 10.1088/0967-3334/28/1/007
11. Ghimenti S, Lomonaco T, Bellagambi FG, Tabucchi S, Onor M, Trivella MG, et al. Comparison of sampling bags for the analysis of volatile organic compounds in breath. J Breath Res. 2015 Dec;9(4):047110. DOI: 10.1088/1752-7155/9/4/047110
12. Djukanović R, Brinkman P, Kolmert J, Gomez C, Schofield J, Brandsma J, et al. Biomarker Predictors of Clinical Efficacy of the Anti-IgE Biologic, Omalizumab, in Severe Asthma in Adults: Results of the SoMOSA Study. Am J Respir Crit Care Med. 2024 Apr 18; DOI: 10.1164/rccm.202310-1730OC